
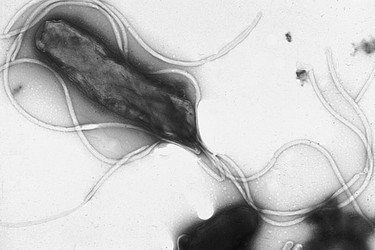
Electron micrograph of Helicobacter pylori possessing multiple flagella (negative staining). Photograph courtesy Yutaka Tsutsumi, M.D. Professor Department of Pathology Fujita Health University School of Medicine.
Introduction
Helicobacter pylori is a species of epsilon proteobacteria which colonizes the harsh environment of the human stomach (Chalmers et al., 2004). Its name refers to both its spiral shape (Helicobacter) and the area of the lower stomach which it habitually colonizes: the gateway (pylorus) between the stomach and small intestine (Meyers, 2007). This bacterium is thought to be present within up to 50% of the human population and has been linked to the development of a number of different medical conditions (Chalmers et al. 2004). This treehouse will provide information about the discovery of H. pylori as well as its classification, morphology, physiology and its effects on its human hosts.
Discovery
H. pylori was first discovered by Robin J. Warren in 1979 at Royal Perth Hospital in Western Australia (Meyers, 2007). Warren had been examining biopsy samples from patients with gastritis (an inflamation of the stomach lining) when he noticed large numbers of curved and spiral bacteria in the biopsy of a human stomach. These bacteria were thriving under the mucus layer of the human stomach and were closely associated with areas of tissue damage. When Warren attempted to present his findings to gastroenterologists at his hospital, he encountered a great deal of indifference because at the time it was thought that no bacteria would be able to survive the harsh environment of the stomach (Meyers, 2007).

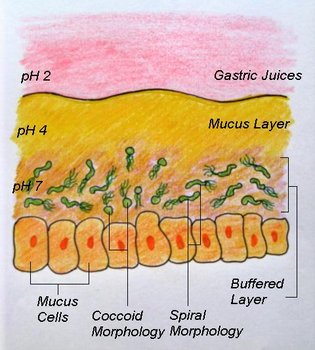
Helicobacter pylori cells under the stomach mucus layer. © Michelle Wiepjes
The first indication of the importance of H. pylori came when Barry Marshall, a resident in internal medicine at Royal Perth Hospital, treated and cured a patient with gastritis using antibiotics (Meyers, 2007). Marshall next attempted to culture the bacteria and succeeded in 1982. Marshall then designed a clinical study and found a very high correlation between the bacteria and certain types of ulcers (Meyers, 2007).
When Marshall and Warren attempted to present their findings to the Gastroenterological community they met with rejection. Even after publication of their joint study in the Lancet in 1983 there was still a high degree of skepticism surrounding the link between bacteria and ulcer formation (Meyers, 2007). In order to prove their theory, Marshall decided that he himself would act as the test subject because lab animals appeared to be immune to the bacteria. After he was proven to have no inflamation or bacteria within his stomach, Marshall ingested some of the cultured H. pylori. After a week he succeeded in giving himself full blown gastritis and proved that the bacteria were responsible for the condition (Meyers, 2007).
By the 1990's H. pylori was not only accepted as the main cause of gastric ulcers, it was also identified as the most common chronic bacterial infection in humans (Meyers, 2007). In 2005 Warren and Marshall were awarded the Nobel Prize for Physiology and Medicine for their discovery and research on Helicobacter pylori.
Classification and Phylogeny
Containing groups: Eubacteria, Proteobacteria, Epsilon-proteobacteria, Campylobacterales, Helicobacteraceae, Helicobacter (Bischoff et al., 2007)

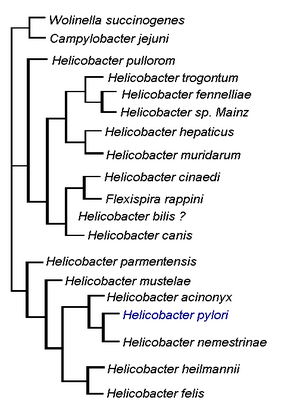
This diagram illustrates the relationship of H. pylori to other members of the Helicobacter genus as well as to members of the Wolinella and Campylobacter genera. © Michelle Wiepjes, created with information from Microbiology of Waterbourne Diseases, 2004.
The bacterium isolated by Warren and Marshall was originally named Campylobacter pyloridis (later changed to the more grammatically correct C. pylori) as it was thought to be a species of Campylobacter (Meyers, 2007). Electron microscopy and genetic studies later revealed that C. pylori has a unique 16S rRNA sequence, sheathed flagella and distinctive outer membrane structures (Meyers, 2007; Chalmers et al., 2004). These structures were sufficiently different from those of Campylobacter that they led to the creation of the new genus Helicobacter. Warren and Marshall's spiral bacteria was then given its current designation of H. pylori (Meyers, 2007).
The Helicobacter pylori Genome
- Genome size: 1.7 million base pairs
- Number of Genes: approximately 1600
- % Genes unique to H. pylori: 17%
Two strains of Helicobacter pylori have genomes which have been completely sequenced (Tomb et al., 1997; Alm et al. 1999). The first genome sequenced was isolated from a gastritis patient in the United Kingdom and was sequenced using the whole genome random sequencing method (Tomb et al., 1997). The second strain of H. pylori was isolated from a patient in the United States in 1994 and was sequenced in 1996 (Alm et al., 1999). The two Helicobacter pylori genomes were compared by Alm et al. (1999) and were found to differ in only six to seven percent of genes. These genes appeared to be clustered within a single region of the genome (Alm et al., 1999).
Morphology
H. pylori is a Gram-negative bacterium which is enclosed within two membranes (Curry & Jones, 1990). When first observed by Warren and Marshall in biopsy samples from gastritis patients, H. pylori was found to have a spiral shape; however this bacterium has also been observed to take on curved, rod (bacillary) or circular (coccoid) forms (Meyers, 2007; Chalmers et al., 2004). H. pylori typically has between four to seven flagella localized at one pole of the bacterium (Salyers & Whitt, 2002). The combination of its curved shape and unipolar flagella allows H. pylori to move easily through the thick mucus layer of the human stomach (Salyers & Whitt, 2002).

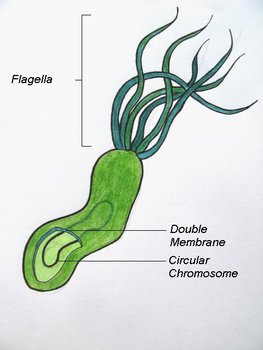
The spiral morphology of Helicobacter pylori © 2008 Michelle Wiepjes
Studies conducted by Ng et al. (1985) found that within colonies of H. pylori, the circular, or coccoid forms were found at the centre of the colony while the spiral forms were found at the edges and were actively dividing. It is theorized that the circular form of H. pylori is inactive and represents a survival adaptation which allows the organism to survive unfavourable conditions (Curry & Jones 1990). It is also theorized that it is the coccal forms that are involved in the transmission of H. pylori.
Physiology
Helicobacter pylori has a microaerophilic physiology meaning that it thrives best in low oxygen enviroments (Helicobacter Foundation, 2006). Analysis of the Helicobacter pylori genome sequenced by Tomb et al. (1997) indicated that glucose was the primary carbon source used for energy production.
Another interesting aspect of H. pylori physiology is that it thrives at a neutral pH of 7.0 (Helicobacter Asssociation, 2006). In order to protect itself from the acidic environment of the stomach, H. pylori burrows its way into the stomach's mucus lining (Helicobacter Asssociation, 2006). There H. pylori use the powerful enzyme urease to maintain a favourable pH by breaking down urea in the stomach into ammonia and bicarbonate–strong bases which counteract the acid in the stomach (Chalmers et al., 2004). Urease enzyme activity has been shown to be essential to the colonization of H. pylori and as a result, H. pylori has developed a unique 2 subunit urease enzyme that is exceptionally powerful (Chalmers et al., 2004).
H. pylori obtains its nutrients by taking advantage of the human inflammatory reponse. The human body will send extra nutrients to the area colonized by H. pylori in order to help white blood cells attack the bacteria (Helicobacter Foundation, 2006). H. pylori however, are inaccessible to these cells because of their location within the mucus and are therefore able to use the excess nutrients provided by their host with no risk to themselves (Helicobacter Foundation, 2006).
H. pylori Infection and Human Disease
Helicobacter pylori infects the lower regions of the mucus layer of the human stomach and is associated with gastritis and the formation of both gastric and duodenal (upper intestinal) ulcers (Helicobacter Foundation, 2006). It has also been linked to the formation of gastric lymphoma and other cancers (Helicobacter Foundation, 2006). Once infected the human host can remain affected for life unless treated with strong antibiotic or antimicrobial therapies (Chalmers et al., 2004). Infected individuals do not necessarily express symptoms, and it is believed that up to 50% of the human population are infected with the bacteria (Chalmers et al., 2004).
H. pylori can be detected through biopsy of the stomach, through a breath test which looks for exhalation of the excess carbon dioxide produced when the bacteria break down urea, or through a blood test which looks for antibodies created when the body's immune system responds to the H. pylor infection (Helicobacter Foundation, 2006).
Although the transmission route of H. pylori is unknown, it appears to be related to the lack of access to clean water (Helicobacter Foundation, 2006). The faecal-oral route, which would transmit H. pylori through contaminated food and water sources, is the most commonly supported theory of H. pylori transmission (Chalmers et al. 2004).


 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site